As both industry and academia look ahead to the next era of transformation in AI/ML, there are many predictions and assumptions about what solutions will emerge. But one thing is indisputable: better data and privacy protection will be foundational to many of those advancements. As our reliance on data grows, the need for data privacy will become even more imperative for businesses.
For Inpher, our focus has been consistently on two fronts: 1. Develop a fully deployed offering that helps companies leverage differentiated data within and across the organizations while protecting data privacy; 2. Invest heavily in R&D to push the boundaries of PETs, including developing newer, efficient privacy-aware ML algorithms and optimizing functionality to support real-world use cases.
To that end, Inpher’s work has been selected by two premier peer-reviewed conferences this week Privacy Enhancing Technologies Symposium 2022 and IEEE Cloud 2022.
XORBoost: Tree Boosting in the MultiParty Settings
Our paper on XORBoost: Tree Boosting in the MultiParty Settings [Kevin Deforth, Marc Desgroseilliers, Nicolas Gama, Mariya Georgieva, Dimitar Jetchev, and Marius Vuille] was accepted at PET Symposium 2022 and its proceedings PoPETs. Gradient boosting is a well-known ML technique used for regression and classification problems. XGBoost is one of the most popular open-source libraries that supports gradient boosting for various programming environments and architectures. In this paper, we present XORBoost, a novel protocol for training gradient boosted tree models and for inferring with these trained models on sensitive data using MultiParty Computation. The data could be horizontally (more features) or vertically stacked (more samples) and stored across silos either within or across the organizations. With XORBoost, the information on features, thresholds, and evaluation paths are private; only tree depth and the number of binary trees are public parameters of the model. For ease of use, XORBoost follows APIs similar to XGBoost’s sklean python API but with security integrated, making it easier for data scientists to leverage sensitive data for building models.
While XORBoost is agnostic to the underlying MPC method or framework, we have used the Manticore MPC framework for its following advantages:
-
Never overflows [an important feature for ML applications] without compromising security or efficiency
-
Keeps the data as modular integers instead of other inefficient methods, converting them to Boolean shares
-
Leverages Manticore’s oblivious sorting and oblivious permutation functionality, thereby improving the performance of XORBoost
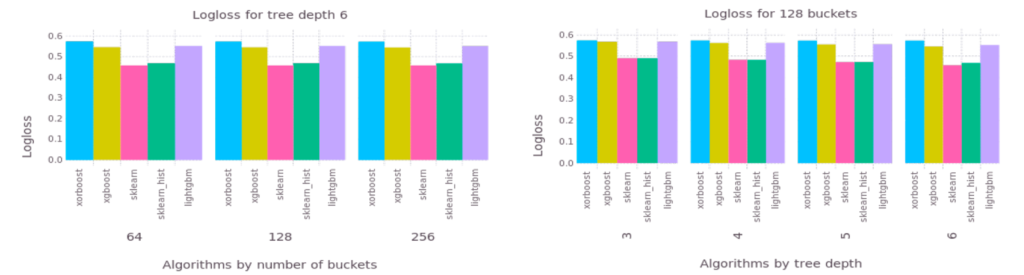
Figure 1: Logloss comparison with plaintext implementation
When we compared XORBoost with several well-known plaintext implementations, minimal predictive power was lost. Read this blog post to learn more about XORBoost and to try it out with a fraud detection use case.
GenoPPML – A Framework for Genomics Privacy-Preserving Machine Learning
Advances in biomedical analytics and AI have revolutionized modern healthcare. Predictive systems in this field allow for better medical and epidemiological research and assist in tailoring proactive healthcare plans that can save lives. In 2020, Inpher’s team participated in the iDASH Privacy and Security Workshop along with other leading research labs and pioneering companies to solve complex ML problems in the privacy-sensitive genomics use cases without leaking any confidential information. The goal was to help genomics researchers diagnose problems early on by building an accurate ML model that leverages individuals’ sensitive genomics data beyond publicly available data such as ENCODE, PDB, TCGA, etc.
Inpher’s team won both the tracks it participated in:
-
Secure Multi-Label Tumor Classification using Homomorphic Encryption
-
Differentially Private Federated Learning for Cancer Prediction
We built a novel framework, named GenoPPML, for an end-to-end privacy-preserving genomics ML pipeline by unifying the above approaches. It uses a logistic regression model over datasets from several private genomics database owners using Multiparty Computation for training. A differential private joint model is revealed to the analyst party. The analyst uses this model to provide prediction services on homomorphically encrypted genomics data samples using TFHE. GenoPPML was submitted by Inpher’s team (Sergiu Carpov, Nicolas Gama, Mariya Georgieva, and Dimitar Jetchev) and accepted in IEEE Cloud 2022.
We benchmarked the solution with three datasets from the TCGA (The Cancer Genome Atlas) database, specifically with genomic data.
Inpher’s team won both the tracks it participated in:
-
BC-TCGA dataset contains gene expression data for 17,814 genes and 590 samples (61 normal tissue vs. 529 breast cancer tissue samples)
-
GSE2034 dataset contains gene expression data for 12,635 genes and 286 samples (107 recurrence tumor samples vs. 179 no recurrence samples)
-
BC12-TCGA dataset contains genomic data for 25,128 genes and 2713 samples for 11 tumor locations
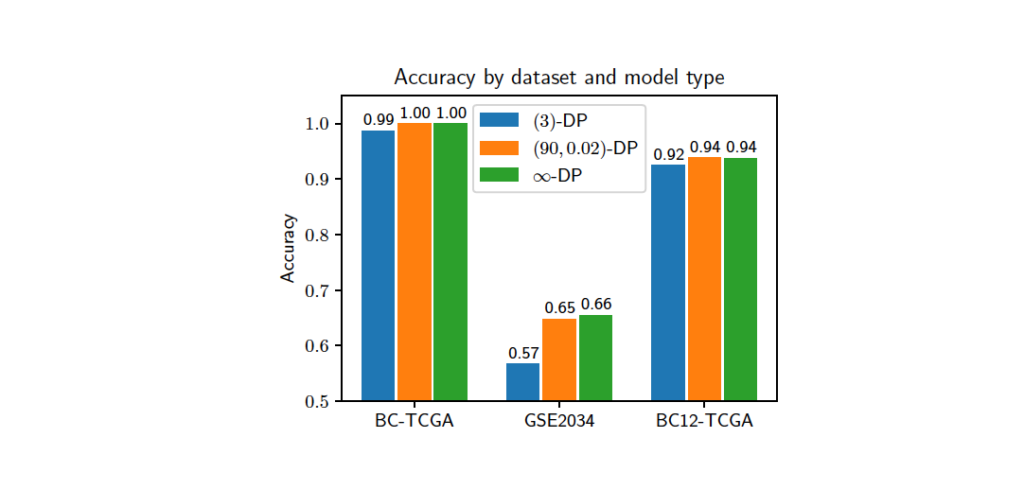 Our objective was to predict if a given tissue sample, described by its gene expression data, belongs to the cancerous or normal group for the BC-TCGA dataset. The privacy budget ε varies from 3 to 90, and δ is either 0 (ε-DP) or 0.02 ((ε, δ)-DP). We also tested the model with no additional DP noise (∞–DP). The image above shows the best model accuracy scores obtained for most secure (3,0)-DP, least secure (90,0.02)-DP, and noiseless ∞–DP settings. The results clearly show that the obtained models achieve state-of-the-art accuracy, and training times are lower when compared to existing works. Additionally, the prediction phase is almost instantaneous even with homomorphically encrypted data, making it suitable for low-latency applications. To learn more on GenoPPML, download the published paper here.
Our objective was to predict if a given tissue sample, described by its gene expression data, belongs to the cancerous or normal group for the BC-TCGA dataset. The privacy budget ε varies from 3 to 90, and δ is either 0 (ε-DP) or 0.02 ((ε, δ)-DP). We also tested the model with no additional DP noise (∞–DP). The image above shows the best model accuracy scores obtained for most secure (3,0)-DP, least secure (90,0.02)-DP, and noiseless ∞–DP settings. The results clearly show that the obtained models achieve state-of-the-art accuracy, and training times are lower when compared to existing works. Additionally, the prediction phase is almost instantaneous even with homomorphically encrypted data, making it suitable for low-latency applications. To learn more on GenoPPML, download the published paper here.

